Creating a Jenkins Data Science Platform
Having worked with Jenkins-CI in the life sciences context for the last few years, I am finding that Jenkins satisfies key requirements of a robust framework for efficient data pipelining integration, and analysis. Here I will briefly describe the attributes that make Jenkins-CI a suitable platform for data scientists
In daily practice, lab and data scientists require tools that allow them to integrate a variety of data management and data analysis tasks. Scientific data flows from a variety of instruments and in a variety of formats. Frequently, data needs to be transformed and visualized for quality control purposes before an analysis can even begin. Experimental annotation and analysis metadata need to be overlaid with the instrument measurements and presented in context for improved comprehension of the results. Finally, the data needs to be processed with domain specific tools (such as image processing, sequence analysis, database searching, statistical learning etc.) to generate high quality results from the raw data 1.

In all of these tasks, having easy to use data pipelining tools that scientists can use to progressively manage, visualize, and analyze their data is critical for the efficient, accurate and reproducible processing of scientific data. Various data tasks and tools need to be used and integrated while maintaining an accurate record of their use and intermediate results. Furthermore, in the spirit of team collaboration (another cornerstone of science), raw, intermediate, and final data need to be shared in a transparent and timely way with others.
Having worked with Jenkins-CI in the life sciences context for the last few years, I am finding that it satisfies many of these requirements and so it can serve in its own right as a robust framework for efficient data pipelining integration, and analysis.
Here I will briefly describe some of the attributes that make Jenkins-CI a suitable platform for data scientists
Data Management
A variety of data can be maintained in the Jenkins artifact archives. Builds can upload, access and efficiently process data from a variety of sources to generate and annotate file artifacts. A variety of plugins support typical data management tasks such as copying, archiving, deleting and moving files across file systems and cloud services. Furthermore, Jenkins provides excellent provenance information and tools for file artifacts (such as logs, date-stamps, and fingerprints).
Although Jenkins is not currently backed by a database system this has the advantage of simplicity and flexibility. In addition, Jenkins provides an extensive API for querying build and artifact information across the entire build system thus providing some of the advantages of structured database storage.
General Data Munging and Processing
This is one of the areas where Jenkins really shines. Any external process or tool can be easily integrated as a build step. Data ‘wrangling and munging’ are important data science competencies that can be carried out with ease on Jenkins. Support for the Groovy dynamic scripting language and Python, which is popular with data scientists, provides an unlimited way of integrating custom scripts and external programs into Jenkins workflows without the need to write custom plugins. Support for ssh allows remote execution of commands and can be easily adapted for high performance parallel computing tasks. Scientific software packages may be available only on certain compute servers. Using ssh build steps you can easily execute these packages on the remote servers and then manage the output using Jenkins. The open source BioUno project has pioneered the implementation of scientific software packages as Jenkins plugins 2.
Integration with Domain specific Software Packages
In the same way that Jenkins can integrate general data ‘wrangling and munging’ tasks, it can also pipeline data through domain-specific software. We have already demonstrated Jenkins applications in areas such as phylogenetics, genetic analysis and image analysis [3,4]. The BioUno project has pioneered the development of domain specific plugins for Jenkins, but in general any software package that can be executed and parametrized from the command line can be easily executed, monitored and pipelined as a Jenkins project. In fact we have found that these domain-specific applications benefit from being ‘wrapped’ as Jenkins projects in that they become easier to access and easier to use by laboratory scientists without the need for dedicated computer savvy data scientists.
Statistical Data Analysis
At this time Jenkins has a limited number of data analysis plugins, but the ones that are available can provide crucial support for data science. For example, the Jenkins R-plugin [5] is simple yet powerful. The R plugin allows you to take full advantage of the R statistical language, the ’lingua-franca’ of scientific data analysis and visualization. Python another popular scripting language for analytics, visualization and machine learning is fully supported in Jenkins through the Python plugin [6] and can also be used for data analysis tasks. Jenkins support for both R and Python make it a compelling tool for data scientists working with modern statistical and machine learning algorithms. Interestingly, using Jenkins to maintain data-science artifacts, creates a supportive environment for reproducible research. The analytical components and results from multiple analyses can be managed, evaluated, compared and validated using testing and verification tools popularized by the software engineering community (such as SCM, testing etc.)
Data Visualization and Communication
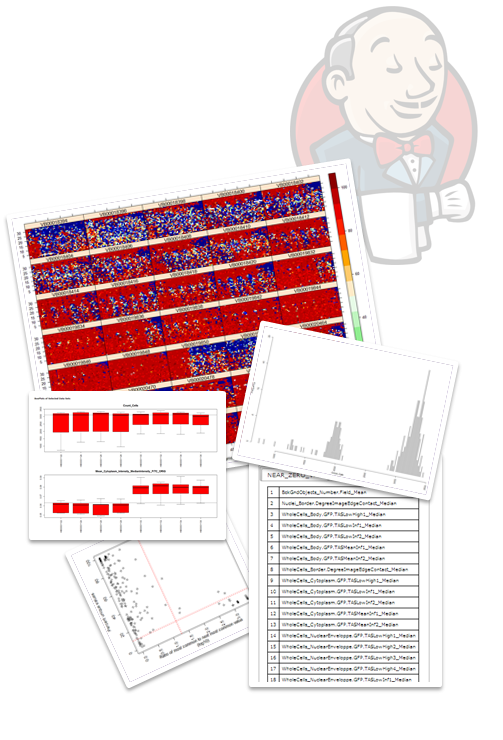 Using Jenkins for data science assumes that we can easily generate, and support graphical visualizations and compelling data reports. R and Python packages can generate a variety of visualizations and graphics. Luckily most of these visualizations are in web standard formats (PNG, JPEG, PDF etc.) that can be compiled into useful reports using existing Jenkins plugins. The [Image Gallery](https://wiki.jenkins.io/display/JENKINS/Image+Gallery+Plugin) [7], [Summary Report](https://wiki.jenkins.io/display/JENKINS/Summary+Display+Plugin) [8] and [HTML Publisher](https://wiki.jenkins.io/display/JENKINS/HTML+Publisher+Plugin) [9] plugins are some of the Jenkins plugins that I have successfully used to create comprehensive graphical reports and visualizations.
Using Jenkins for data science assumes that we can easily generate, and support graphical visualizations and compelling data reports. R and Python packages can generate a variety of visualizations and graphics. Luckily most of these visualizations are in web standard formats (PNG, JPEG, PDF etc.) that can be compiled into useful reports using existing Jenkins plugins. The [Image Gallery](https://wiki.jenkins.io/display/JENKINS/Image+Gallery+Plugin) [7], [Summary Report](https://wiki.jenkins.io/display/JENKINS/Summary+Display+Plugin) [8] and [HTML Publisher](https://wiki.jenkins.io/display/JENKINS/HTML+Publisher+Plugin) [9] plugins are some of the Jenkins plugins that I have successfully used to create comprehensive graphical reports and visualizations.Finally, results can be easily communicated between data and laboratory scientists through Jenkins and the build in access and authorization tools that the platform provides
Summary
In summary, I feel that Jenkins makes an excellent platform for data science experimentation and for providing practical and easy access to data science algorithms and visualizations to lab scientists. Although several challenges remain for making Jenkins a mainstream platform for data scientists (many are currently experimenting with the concept of analysis ’notebooks’, such as iPython, Apache Jupyter, Beaker [10-12] etc.), I content that an even more significant challenge stems from the historical user base of Jenkins. I have found that many DevOp engineers are ambivalent about the life/data-science uses of Jenkins that I and my colleagues at the BioUno project are proposing. Although many appreciate the demonstrable power that Jenkins can inject into computational life/data sciences, many worry about the complexities required to support Jenkins in these challenging, demanding and less understood (from a DevOp engineer’s view point) research areas. Many DevOp shops want to have control over the Jenkins configuration, plugins and environment, and rightly so, don’t want a data scientist to dictate additional complexity. Even so, I have found that the ease with which one can deploy a working instance of Jenkins and effectively use it outside the DevOp Jenkins environment makes Jenkins a relatively easy addition to the toolset that data scientists can deploy and maintain on their own. Of course, it would be great if the DevOps community was to accept and contribute to these new and open research areas, and I hope that the BioUno project will become one of many connecting nodes between DevOp engineers and life/data scientists.
- http://blog.udacity.com/2014/11/data-science-job-skills.html
- http://biouno.org/
- http://biouno.org/jenkins-plugins.html
- https://figshare.com/articles/Jenkins_as_a_Scientific_Data_and_Image_Processing_Platform/1098763
- https://wiki.jenkins.io/display/JENKINS/R+Plugin
- https://wiki.jenkins.io/display/JENKINS/Python+Plugin
- https://wiki.jenkins.io/display/JENKINS/Image+Gallery+Plugin
- https://wiki.jenkins.io/display/JENKINS/Summary+Display+Plugin
- https://wiki.jenkins.io/display/JENKINS/HTML+Publisher+Plugin
- http://ipython.org/notebook.html
- http://jupyter.org/
- http://beakernotebook.com/