rOpenSci in Jenkins
rOpenSci is an Open Data project. It has many R packages that access several data repositories. In this post we will demonstrate how to use rOpenSci taxize package with Jenkins and the R Plug-in.
Install necessary tools
You will have to install the following tools in your computer in order to run this example.
- Java
- Jenkins
- R Plug-in for Jenkins
- Image Gallery Plug-in for Jenkins
- R
- taxize
We won’t go into detail here about how to install each tool. You will need Java for running Jenkins, R for running the example and the taxize R package. The R Plug-in will allow Jenkins to run R scripts and the Image Gallery will create a simple gallery using the image created by the build in Jenkins.
Setting up the Jenkins job
Once you have the plug-ins configured and Jenkins running, create a FreeStyle job and give it any name you want.
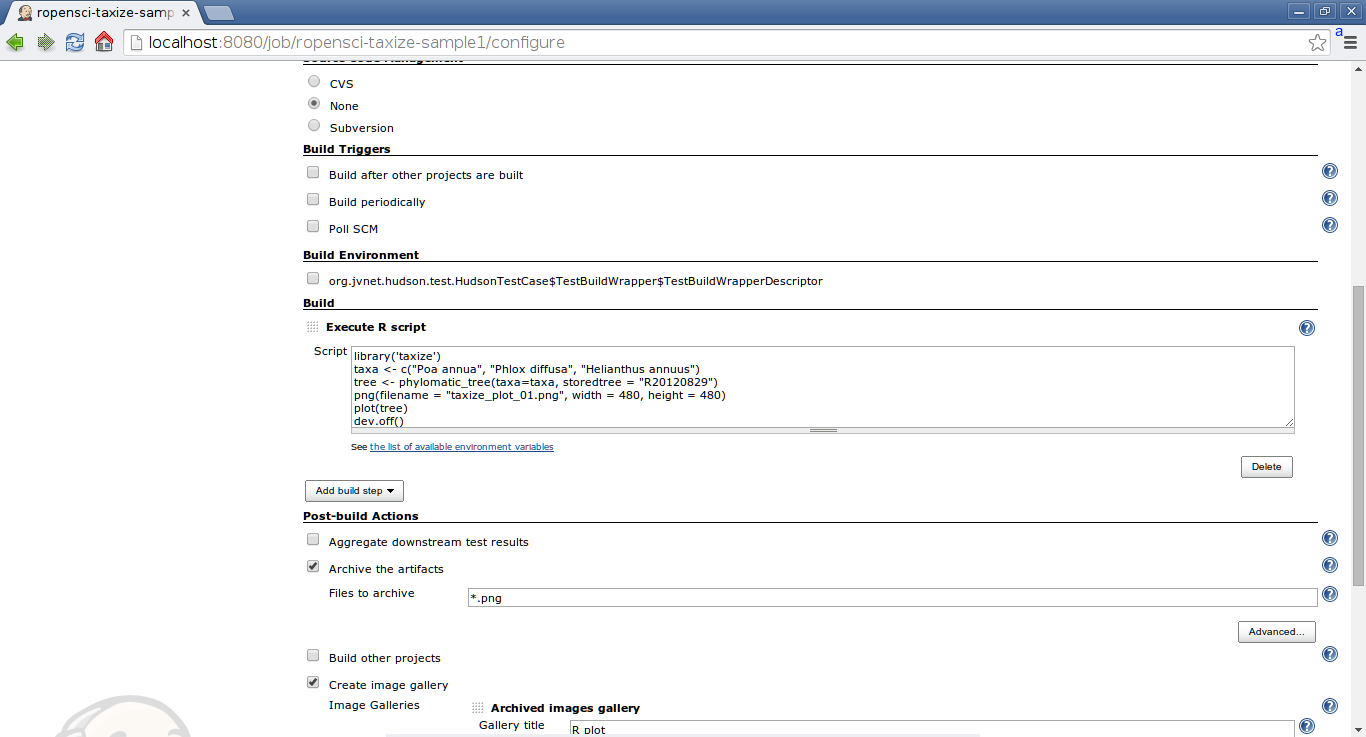
Now click the “Add build step” and select “Execute R script”. That will add a textarea where you can write your R script. Let’s copy and paste the taxize example from rOpenSci, with one modification to create a PNG with the tree.
{% highlight r %} library(’taxize’) taxa <- c(“Poa annua”, “Phlox diffusa”, “Helianthus annuus”) tree <- phylomatic_tree(taxa=taxa, storedtree = “R20120829”) png(filename = “taxize_plot_01.png”, width = 480, height = 480) plot(tree) dev.off() {% endhighlight %}
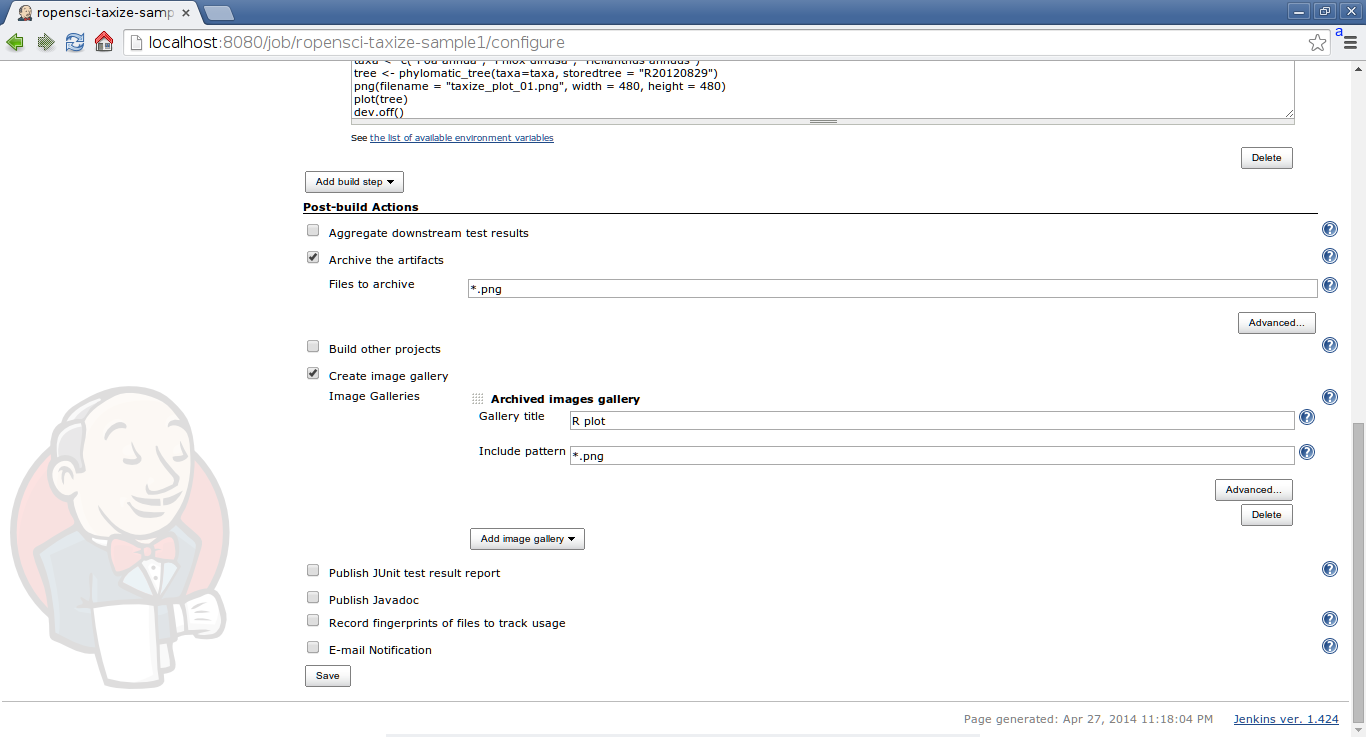
The next step is the last before you can build your project. Your job is already configured to run the taxize example, but let’s add an image gallery as post build step. In order to do that we have to archive the PNG created by our R script.
Check the box that says “Archive the artifacts” and use the pattern *.png. Also check the box that says “Create image gallery” and add an “Archived images gallery”, again using the *.png regex pattern and give it any name.
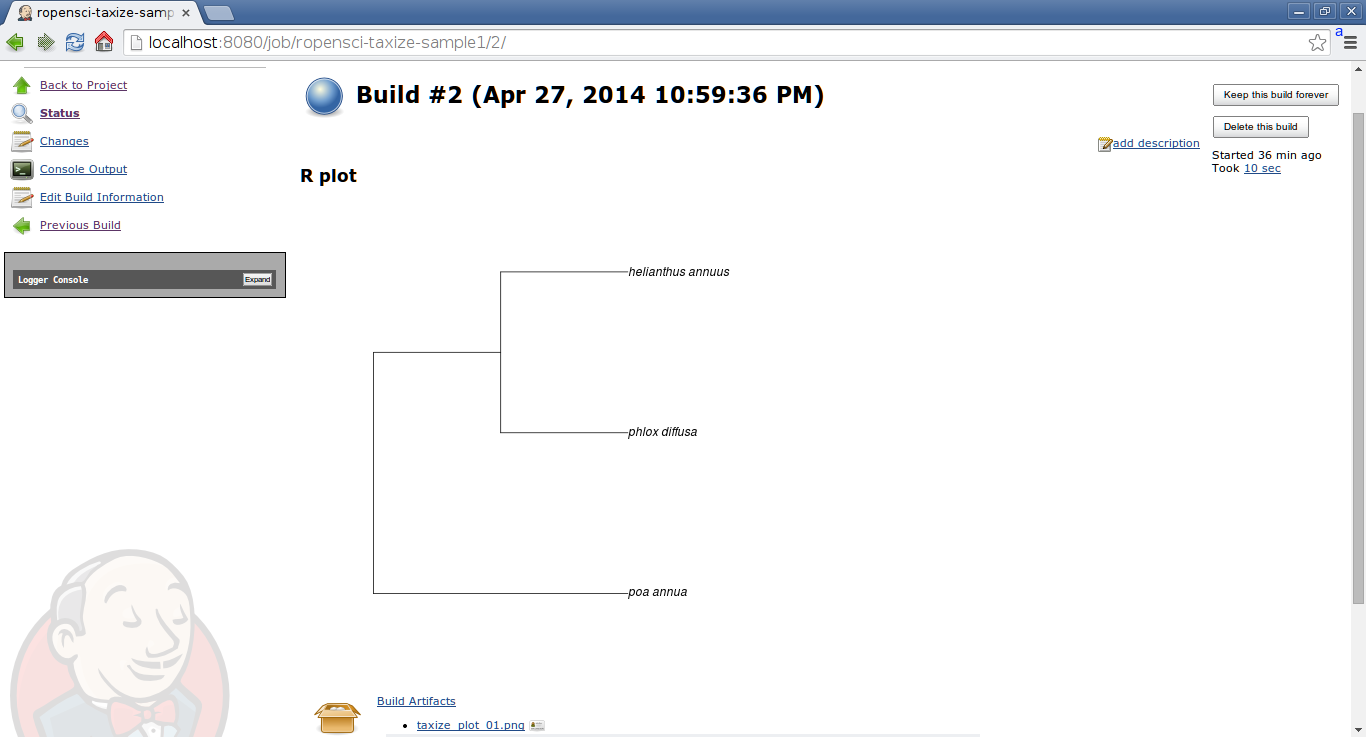
Build your job and check the results in the console log. You will also find the image being displayed in the build page.
Benefits of using rOpenSci and Jenkins
While you can use rOpenSci without Jenkins, here are some benefits of not doing so:
- You can add a Jenkins slave that will execute your scripts in another computer. This is specially useful if you have R installed in another machine.
- Jenkins will store the console output of each build for you
- You can trigger jobs in Jenkins via your browser, Android/Apple device, or using web hooks like with GitHub commits
- Your artifacts will be archived, meaning that you can use them in other jobs and back it up too.
- Jenkins has security control built in, as well as notifications (e-mail, SMS, Google Talk)
Likewise you can use Jenkins and R without rOpenSci, but then you have to create another way to retrieve the data. Even if you used the same datasource that rOpenSci uses, you would have to write tests to assert that you don’t have bugs in your code.
rOpenSci packages are well written, and are being tested by many users and used in publications already. So if you didn’t know rOpenSci, go check it out. If you already did, what are you waiting to try Jenkins and rOpenSci together :-)